Modelling metabolic pathways of an organism plays an important role in elucidating phenotypic changes as well as genotype-phenotype relationships.
Kinetic simulations using Ordinary Differential Equations (ODEs) allow us to model the dynamic behaviour of the cell as enviromental systems change.
Kinetic simulations using Ordinary Differential Equations (ODEs) allow us to model the dynamic behaviour of the cell as enviromental systems change.
|
Given a complex set of kinetic reactions as shown in Figure 1, parameter estimation becomes important to determine the approximate values of the kinetic parameters.
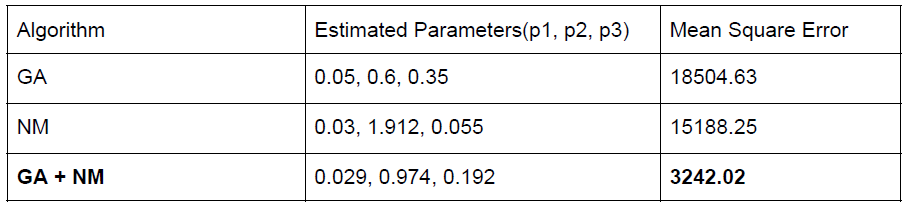
In this project, I explore a combination of local and global optimization to find accurate estimates of kinetic parameters for an MAPK cascade. Specifically, I use a Genetic Algorithm for global parameter search, followed by Nelder Mead simplex for local search. I also compare this with using only local or only global search heuristics, and show that the combined algorithm gives more accurate estimates, as evinced by the graphs and table in Figure 2 below. The code for this is available on my Github. |
The table in Figure 2 shows the comparison of using the combined optimization as against individual heuristics in terms of total error using the estimated parameters, underlying the effectiveness of using Genetic algorithms as a global search heuristic followed by Nelder-Mead for local search.
Figure 2: Parameter estimation using (1) Only Nelder-Mead Simplex [NM] (left), (2) Using only genetic algorithm [GA] (middle), and (3) Using Nelder-Mead initialized using genetic algorithm estimates (right). Simulated ODE solution at the top in each image, experimental observed concentration below