Related Publications:
Zichen Wang, Parth Natekar, et al. "MitoTNT: Mitochondrial Temporal Network Tracking for 4D live-cell fluorescence microscopy data." bioRxiv 2022.08.16.504049 (Link).
Zichen Wang, Parth Natekar, et al. "MitoTNT: Mitochondrial Temporal Network Tracking for 4D live-cell fluorescence microscopy data." bioRxiv 2022.08.16.504049 (Link).
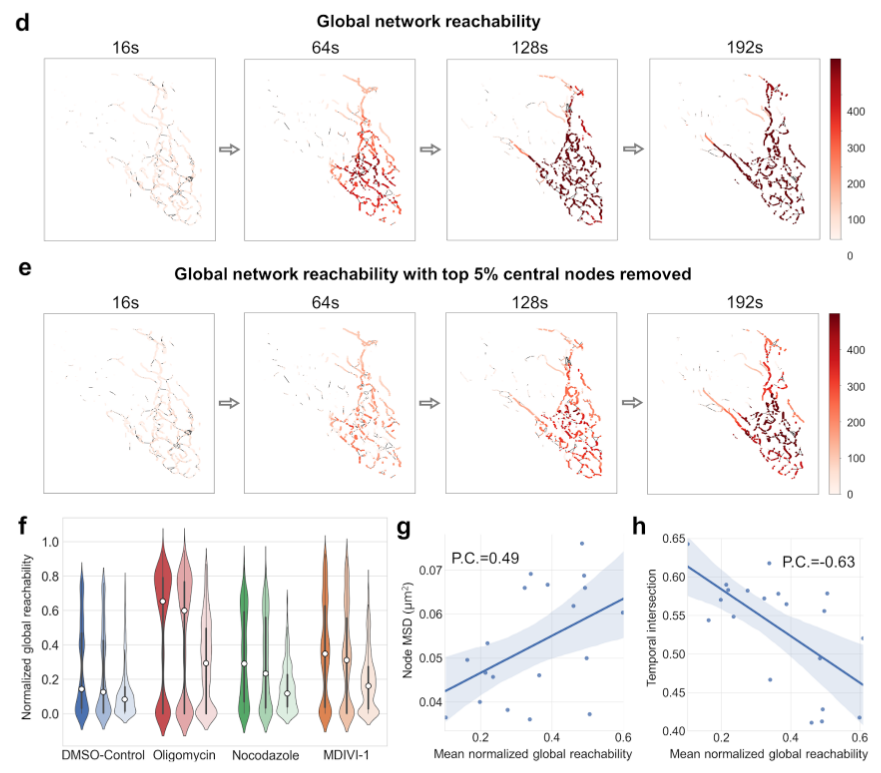
Lattice-light sheet microscopy has recently enabled extracting 4D (3D + time) volumes of cell images. It is thus possible to model Mitochondria as 4D networks, i.e 3D temporal networks. We leverage this to look at mitochondrial behaviour and dynamics at a previously unprecedented level, demonstrating, among other things, how graph-based methods can be leveraged to analyse these 4D mitochondrial networks to reveal cross-network transport patterns and responses to simulated pharmocological interventions.
|
The figure to the right is an example of how temporal graph modeling can answer questions about network properties on a sub-cellular level. It shows how global network reachability is affected when 5% of the most central nodes are removed, the relative effect of removing 5% random nodes, and the correlation between global reachability and temporal graph metrics. One could then ask about how network reachability is affected in diseased cells, and what pharmacological interventions would bring the network back to a healthy state. |